Annals of Neurosciences, Vol 17, No 2 (2010)
Annals of Neurosciences, Volume 17, Issue 2 (April), 2010
Phylogenetic analysis of poly and non structural protein in Japanese Encephalitis virus with other related viral families
KEY WORDS
Multiple Sequence Alignment
Phylogenetic Analysis
Japanese Encephalitis
Corresponding Author
Prachi Srivastava, Ph.D
Tel. : +91-9453141916
E-mail : p.srivastava@amity.edu
prachi767@rediffmail.com
ABSTRACT
Background : Japanese encephalitis (JE) causes inflammation of brain. The mortality rate due to JE is 30% while 10 –15 % of patients make full recovery. The disease spreads through infected mosquito bites breeding in rice fields and feeds on pigs, birds, and ducks. Purpose : As proteins show important structure to function relationship the study was designed to carry out the identification of poly and non-structural proteins in the infective virus group using different strains of Japanese encephalitis virus i.e. JAOARS982, Nakayama Strain SA (V), Strain SA-14. Methods: With reference to non structural proteins we obtained protein sequences of the following Japanese encephalitis virus groups: Japanese encephalitis virus, Weatnile virus, Kunjin virus. Further comparative and phylogenetic analysis was performed to explore evolutionary relationship among these groups. Results: Results of phylogeny of alignment score was found to be 375184 using multiple alignment, Jal view, ClustalW (1.83) and ClustalW2. However, the analysis among the non-structural proteins of Japanese Encephalitis Virus, Westnile Virus, and Kunjin Virus revealed the phylogeny alignment score to be 875 through multiple sequence alignment and Tree view respectively. Conclusion : Phylogenetic analysis revealed that these four strains are interrelated as well as showing high similarity with the other viruses of this group due to conserved regions among their sequences.
doi : 10.5214/ans.0972-7531.1017205
Introduction
Encephalitis is a medical emergency and refers to inflammation of the brain tissue; this can also involve the membranes of the brain. Japanese Encephalitis is a disease caused by the mosquito borne Japanese Encephalitis (JE) Virus. JE is caused by the virus belonging to the family Flavivirida, whose members include several pathogens of humans and animals. Japanese encephalitis virus (JEV) is the cause of the most prevalent viral encephalitis of man in terms of morbidity and mortality.1Approximately 50,000 cases of JE occur annually in Asia. In India, epidemics of JEV have been reported since the mid-1950s, and the virus is now endemic in most parts of the country. This virus is a part of a group of viruses known as arbo viruses that are spread by arthropods (mosquitoes or ticks).2,3 JEV is a mosquito-borne virus which is potentially fatal in humans. Domestic pigs and wild birds are reservoirs of the virus and transmission to humans may cause severe symptoms. One of the most important vectors of this disease is the mosquito Culex tritaeniorhynchus. This disease is spread throughout Eastern Asia including India, Japan, China and Southeast Asia. Structure of the protein has a major role in determining its functions as the active and the regulatory site all are related to the structure of protein. Poly proteins and non structural proteins are important proteins found in JE Virus causing brain fever in human beings. The poly protein (chain A) is a 430 amino acid residue long protein.4 Structure of both (poly and non structural) protein is still unknown. This disease is devastating and causes inflammation of the brain (encephalitis) which can lead to death or brain damage. There is 30% mortality rate for people who get sick from Japanese Encephalitis and only 10-15% of patients make a full recovery.5 Only 1 in every 30-300 people bitten by an infected mosquito becomes infected. Of those who develop symptoms, the disease is fatal in around 25%, usually occurring within the first 10 days.6,7 Among those who survive, up to 50% are left with brain damage. Japanese encephalitis (JE) is a mosquito-borne arboviral disease of major public health importance in Asia.8,9 More than 35,000 cases and 10,000 deaths are reported annually from the region but official reports undoubtedly underestimate the true number of cases.10,11
Our objective is two-fold. First, to identify the poly and non-structural protein in the member of Japanese encephalitis virus group alongwith four strains of Japanese encephalitis virus i.e. JAOARS982, Nakayama. Strain SA (V), Strain SA-14. In poly protein group we retrieved protein sequences of these strain: -POLG_JAEVJ, POLG_JAEVN, POLG_JAEV5, POLG_JAEV1, POLG_MVEV5, POLG_STEVM, POLG_KUNJM, POLG_WNV genome poly protein. We have determined and analyzed the complete protein sequence of poly and non-structural proteins from sequence databases.
Second objective includes phylogenetic analysis of JEV virus group along with four strains of Japanese encephalitis virus for poly and non-structural groups. On the basis of phylogenetic analysis and sequence comparisons some conserved regions among poly and non-poly structural protein sequences were identified. This study reveals that these proteins are closely related and the JEV shows strong similarity with JE Virus group.
Methods
Selection of poly and non structural proteins
The identification of two types of proteins and their strains poly (POLG_JAEVJ, POLG_JAEVN, POLG_JAEV5, POLG_JAEV1, POLG_MVEV5, POLG_STEVM, POLG_KUNJM, POLG_WNV) and non structural proteins (Japanese encephalitis virus, Westnile virus, and Kunjin virus) were retrieved with the help of different main protein sequence repository databases i.e. NCBI, SWISS PROT and UNIPROT. Further, for phylogenetic analysis we used Tree view, ClustalW, ClustalX, and Jalview tools.12,13
Phylogenetic analysis
Phylogenetic motifs are multiple sequence alignment regions that conserve the overall phylogeny of the complete family. Through the comparison to protein strain of JE Virus group it is shown that Phylogenetic motifs represent good predictions. We deduced amino acid sequences to reconstruct Phylogenetic tree (Cladogram and Phylogram). The trees were obtained using neighbour-joining method (NJ) and average distance method by taking BLOSUM62PID standard with JAL View (http://taxonomy. zoology.gla.ac.uk./rod/html CLUSTAL W, (http://ncbi.nih.nlm.gov./clustalw) Jal view tool.14
Results
Proteins are fundamental components of life. They exhibit an enormous amount of chemical and structural diversity enabling them to carry out an extraordinary diverse range of biological functions. The critical feature of a protein is its ability to adopt the right shape for carrying out a particular function. For the analysis two types of proteins: - Poly Protein and Non Structural Protein for Japanese encephalitis virus group are conducted through phylogenetic analysis. The phylogenetic relationships are predicted by using ClustalW (fig 1a, fig 2a) and JAL View (fig 1b, c, d and fig 2b, c, d) software. Clustal W results predict sequence alignment score among these virus groups&' for poly protein and non-structural poly proteins sequences.
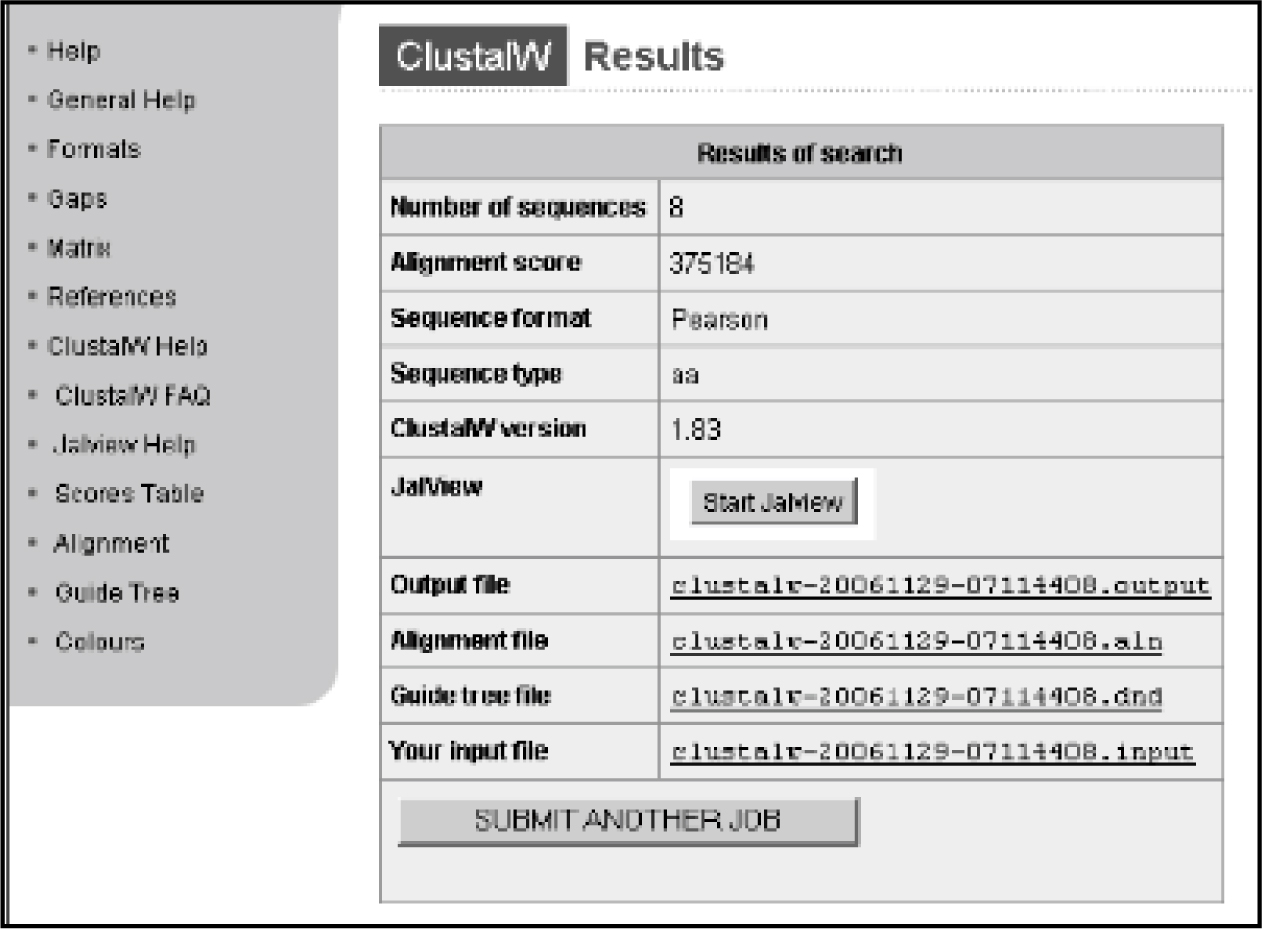
Figure 1 (a): ClustalW Results of Poly Protein
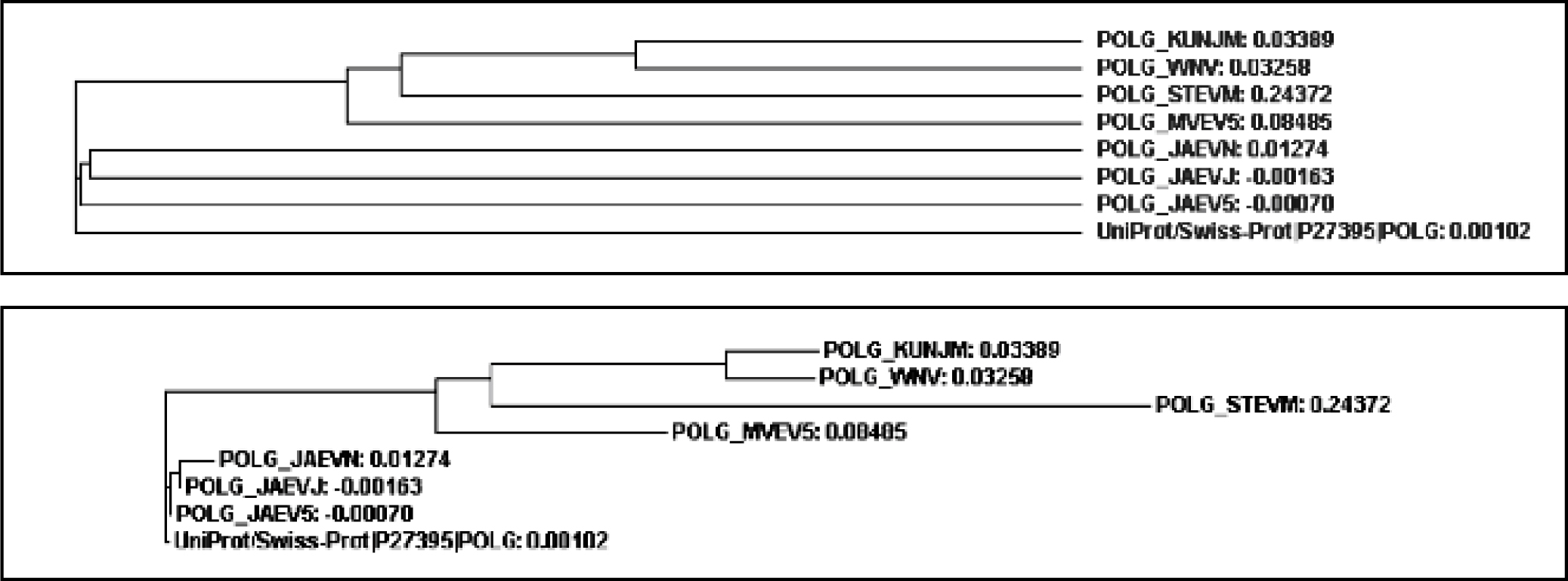
Figure 1 (b): Cladogram and Phylogram tree of Poly Protein
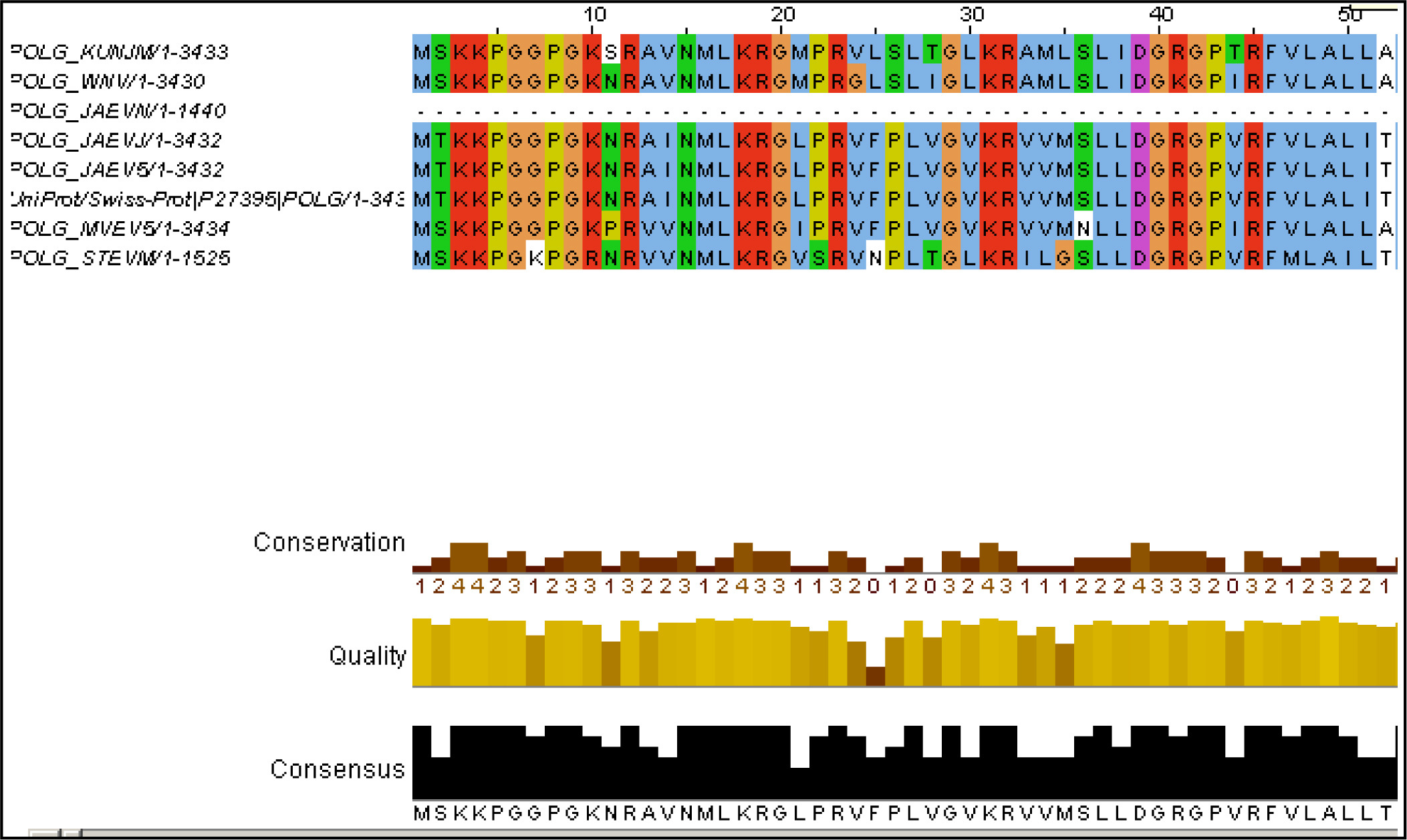
Figure 1 (c): JalView alignment result of Poly Protein
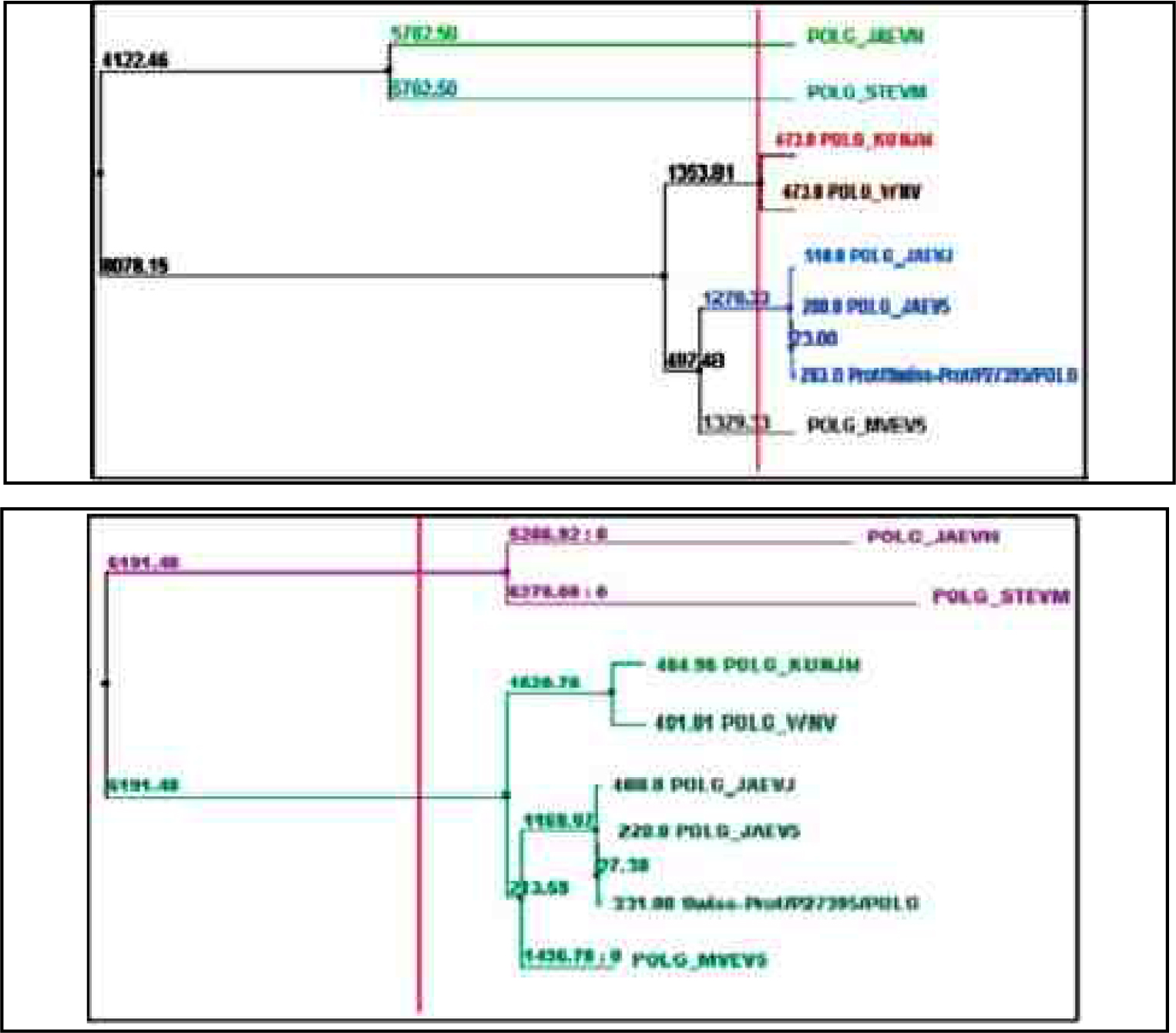
Figure 1 (d): Average distance and Neighbour joining tree using BLOSUM62PID of Poly Protein
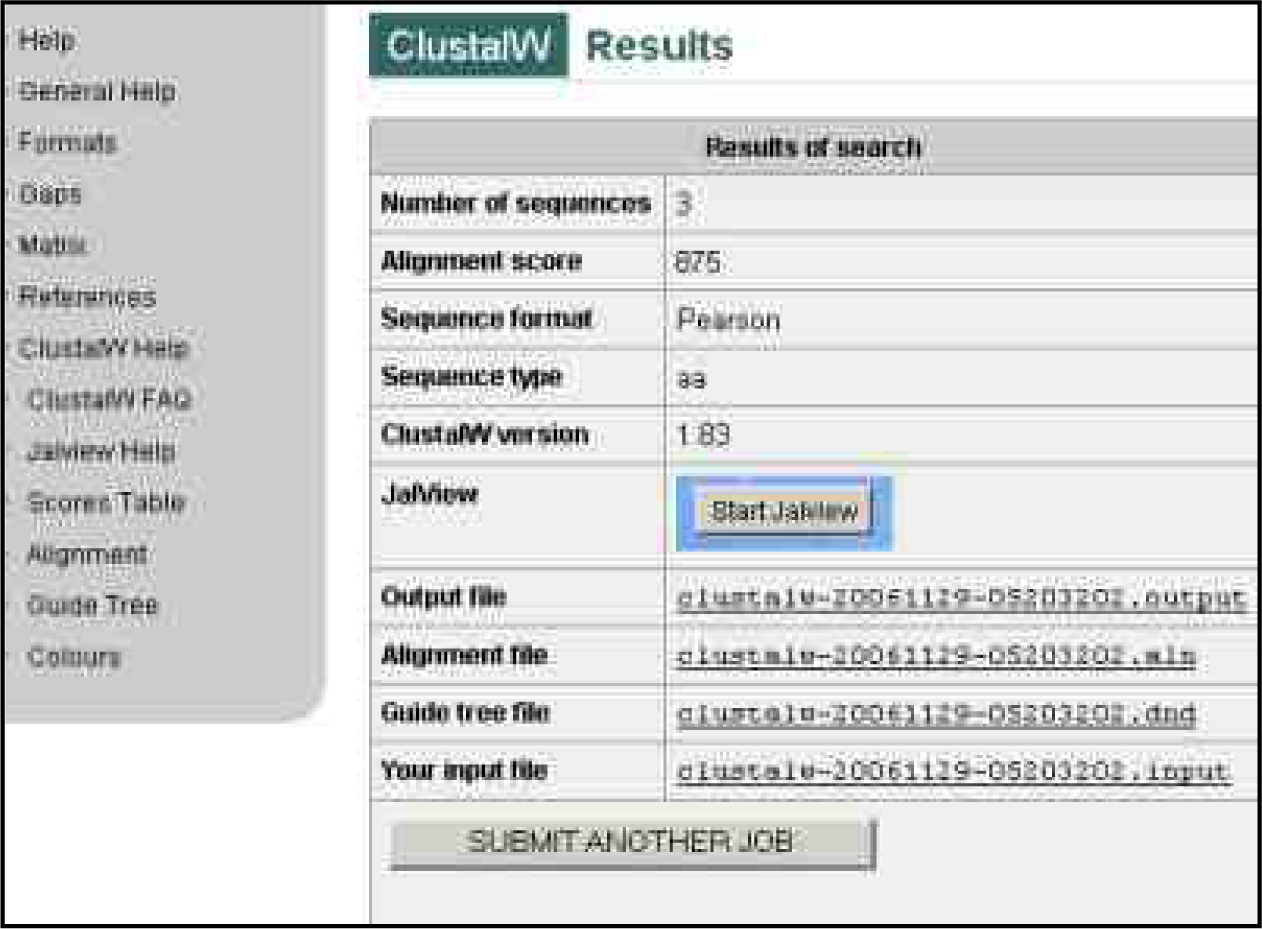
Figure 2 (a): ClustalW Result of Non Structural Protein

Figure 2 (b): Cladogram and Phylogram tree of Non Structural Protein
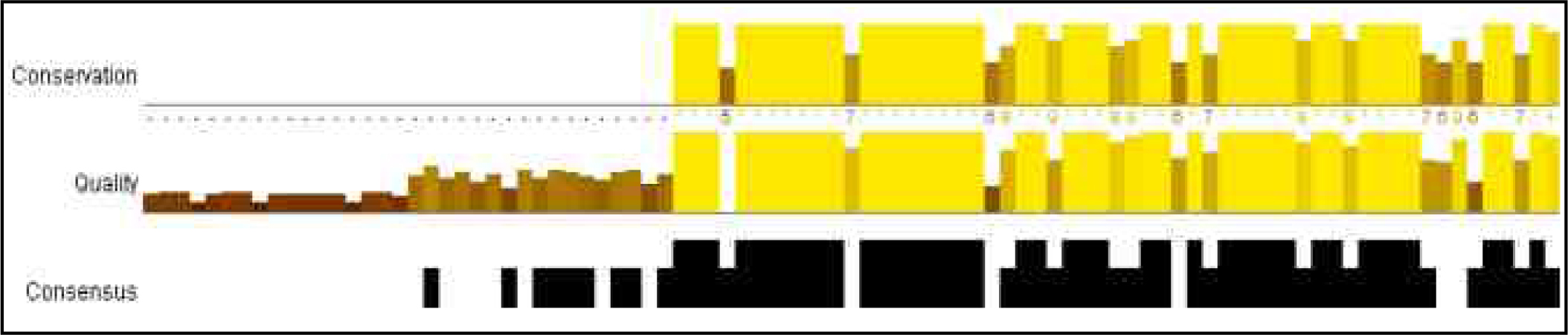
Figure 2 (c): JalView alignment result of Non Structural Protein
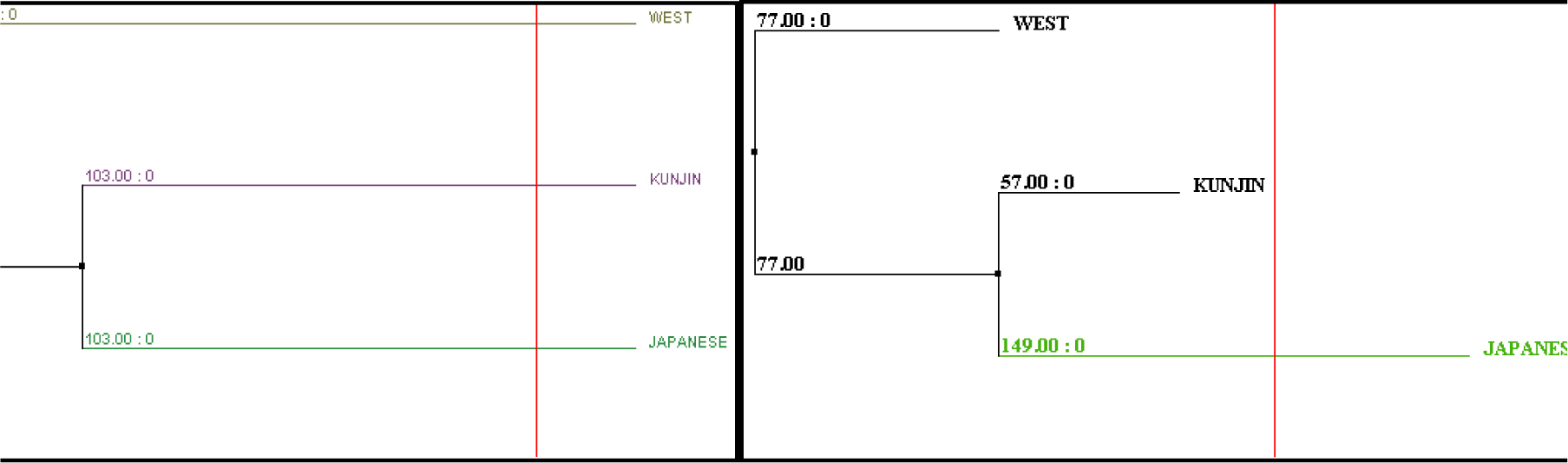
Figure 2 (d): Average Distance and Neighbour joining using BLOSUM62PID of Non Structural Protein
Discussion
Current studies reveal that these four strains are interrelated as well as showing high similarity with the other viruses of this group due to conserved regions among their sequences. The purpose of phylogeny is to reconstruct the history of life and explain the present diversity of living creatures. This can be represented as a huge genealogic tree (the tree of life). The underlying principle of phylogeny is to try to group living creatures according to their level of similarity. Phylogenetics is a special kind of phylogeny that relies on the comparison of equivalent genes coming from several species for reconstructing the genealogic tree of these species and finding out who is the closest relative of whom in the family.
Phylogenetic analysis of protein sequences requires, as a first step, the alignment of amino acid or the corresponding amino acid sequences in such a way that they are homologous in every position. However, not all alignments of homologous sequences are useful for phylogenetic reconstruction, because to make a reliable Phylogenetic tree, sequences should be neither so similar that they are devoid of Phylogenetic information nor so divergent that many positions are saturated by multiple substitution.15
For the phylogenetic analysis of poly and non structural protein constituting Japanese Encephalitis Virus with other related groups we have used certain bioinformatics approaches like Phylogenetic analysis, sequence alignment as well as some phylogenetic analysis tools such as ClustalW & Tree View., A surprising score of (375184) for poly and non structural protein (875) sequences by ClustalW fig1 (a) software which is use for multiple sequence alignment. Further, the cladogram and phylogram for both proteins (poly protein and non structural protein) are generated. For poly protein cladogram and phylogram Fig1 (b) it showed high level of similarity among there tree branches. Therefore, on the basis of this evidence, we predict that POLG_JAEVJ, POLG_JAEVN, POLG_JAEV5, POLG_JAEV1, POLG_MVEV5, POLG_STEVM, POLG_KUNJM, POLG_WNV appears to be evolutionarly related among group of poly protein. Further, for validation we also construct the tree using average distance method and calculated fig 1(e). The tree branches show the boot strap value. All boot strap values show minimum divergence in branches of tree. On the basis of both statistical analysis we predict that these strains: POLG_JAEVJ, POLG_JAEVN, POLG_JAEV5, POLG_JAEV1, POLG_MVEV5, POLG_STEVM, POLG_KUNJM, POLG_WNV group of poly protein show high level of similarity. This is evident from results that these groups of virus strain (Poly protein) are showing high degree of evolutionary similarity.
Second, we performed the analyses as same for non structural protein virus group: Japanese encephalitis virus, Weatnile virus, Kunjin virus. The cladogram and dendogram (fig. 2b) was generated for finding evolution relationship among this group for non-structural protein. On the basis of sequence comparisons by jalview software (fig. 2 c) we identify a highly conserved region among these virus groups (non structural proteins). Then, we validated our results by constructing the tree from average distance and neighbor joining method using BLOSUM62PID (fig. 2 d). We compared there boot strap values showing on branches of trees. These boot strap values also show the minimum divergence. Further, we can conclude that these virus groups of non structural protein (Japanese encephalitis virus, Weatnile virus, Kunjin virus) are evolutionarily closely related and they evolve from same ancestral roots.
Acknowledgements
This is not just to follow the custom of writing acknowledgement but to express and record our heart felt feeling of gratefulness to all those who directly or indirectly helped us in this work. Further we wish to acknowledge Phylogenetic analysis tools for conducting this study.
Competing interests – None, Source of Funding - None
Received Date : 30 April 2010; Revised Date : 26 May 2010
Accepted Date : 28 July 2010
References
1. Service M.W., Agricultural development and arthropod-borne disease - a review. Revista Saude Publicationj 1991; 25:165-178.
2. Gourie-Devi M, Ravi V, Shankar SK. Japanese encephalitis: an overview. Rose FC, ed. Recent Advances in Tropical Neurology. Amsterdam, Netherlands: Elsevier Science Publishers B. V. 1995; 217–235.
3. Patel S, Sinha A, Parmar D, et al. An update on the role of environmental factors in parkinson&'s disease. Annals of Neuroscinces 2006 ; 12: 79-86.
4. Burke DS, Leake CJ. Japanese encephalitis, IN: Monath TP Edthearboviruses: epidemiology and ecology. CRC press, Boca Raton, FL. 1988; 3:63-92.
5. Umenai T, Krzysko R, Bektimirov TA, Assaad FA. Japanese encephalitis: current worldwide status. Bull WHO. 1985; 63:625-631.
6. Nicolosi A, Hauser VA, Beghi E, Kurland LT. Epidemiology of central nervous system infection in Olmsted County, Minnesota, 1950-1981. J Infect Dis. 1986; 154:399-408.
7. Hiroyama T. Epidemiology of Japanese encephalitis (in Japanese) Sai Ching Iga Ku. 1951; 17:1272-1280.
8. Kono R., Ho Kim H. Comparative epidemiological features of Japanese encephalitis in Republic of Korea, China (Taiwan) and Japan. Bull WHO. 1969; 40:263-277.
9. Hullinghorst RL, Burns KF, Choi YT, et al. Japanese B encephalitis in Korea. JAMA. 145:460-466.
10. Sabin AB, Schlesinger RW, Ginder DR., Clinically apparent and in apparent infection with Japanese B encephalitis virus in Shanghai and Tientsin. Proc Soc Exp Biol Med. (1947).65:183-187.
11. Carey DE, Myers RM, Reuben R, et al. Japanese encephalitis in South India. A summary of recent knowledge. J Indian Med Assoc. (1969).52:10-15.
12. Halstead SB., Arboviruses of the Pacific and Southeast Asia, IN: Feigin RD and Cherry JD eds, Textbook of Pediatric Infectious Diseases, WB Saunders, Phila, PA. 1502-1526. Halstead SB, Grosz CR., Subclinical Japanese encephalitis. I. Infection of Americans with limited residence in Korea. Am J Hyg. 1987; 75:190-201
13. Goldman, N. Phylogenetic information and experimental design in molecular systematics. Proc. R. Soc. Lond. B Biol. Sci. 1998; 265:1779–1786.
14. Nei M, Phylogenetic analysis in molecular evolutionary genetics. Annu Rev Gen 1996; 30: 371–403.
15. Thompson JD, Higgins DG, Gibson TJ, et al. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 1994; 22: 4673–4680.
(c) Annals of Neurosciences.All Rights Reserved
